
Most of us give but a passing thought to our mass unless we are dieting or measuring the growth of a baby. Our mass, or weight, is simply a piece of information about us; one that may not be very descriptive without accompanying details of height and coloring.
However, for the invisible molecules that constitute our world, mass can be an incredibly descriptive quality, since the precise mass of a molecule reflects its atomic composition. Analytical chemists measure the mass of molecules by creating gaseous charged ions and recording the mass-to-charge ratio of these ions in an instrument called a mass spectrometer. This ratio can be processed to extract the mass of individual ions. Mass spectrometry can also be coupled with other analytical methods to determine the connectivity between atoms in the molecule.
I have the privilege of conducting my postdoctoral research in the Sharon lab at the Weizmann Institute of Science, in which a specialized type of mass spectrometry is applied to analyze proteins, the large molecules that are the workhorses of the cell. This type of mass spectrometry, called native mass spectrometry (native-MS), creates gas phase ions of whole, intact proteins. We can also create ions of protein complexes that are themselves composed of multiple individual proteins, or of proteins bound to drugs or other ligands. The highly specialized instruments in the Sharon lab have the resolution to distinguish between changes in mass - less than 1% of the overall mass of the protein - that occur upon important biochemical modifications or binding events.
Native-MS is unique amongst bioanalytical tools because it can distinguish between protein modifications or complexes that appear the same to other biochemical techniques. However, if these different forms differ even slightly in their masses, then native-MS can identify and quantify these forms as a function of time or varying sample conditions. This is useful for analyzing how genetic mutations or changes in cellular conditions affect proteins, or the order and tightness with which small molecules bind to protein complexes. As a postdoctoral researcher in the lab, I work to apply native-MS to new types of biochemical problems, as well as to developing new methods and protocols for native-MS.
One such project works on speeding up the applicability of native-MS by bypassing tedious aspects of the sample preparation protocol. For native-MS studies of a protein - and indeed, for most experiments to characterize proteins - researchers have to purify the protein of interest. They start from a sample of cells and end up with a solution containing a single type of protein molecule. However, in the Sharon lab, we work to develop methods and protocols for native-MS of proteins directly from cell homogenates. These methods, dubbed direct-MS, enable users to go from a sample of cells to a mass spectrum in half an hour instead of multiple days of purification.
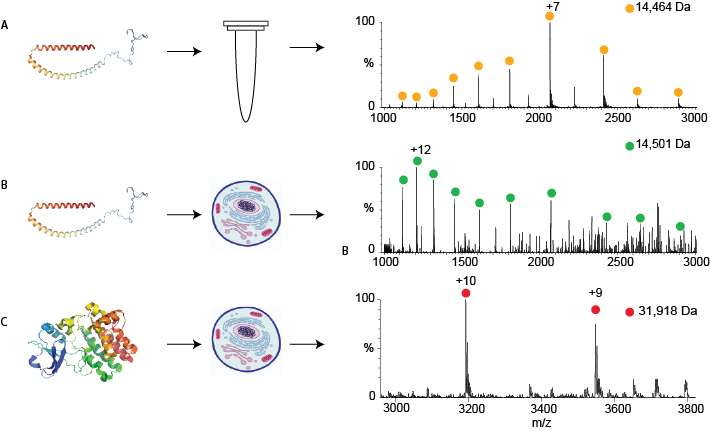
Figure 1:
A- A mass spectrum of a particular protein, alpha-synuclein, which was purified. The peaks corresponding to alpha-synuclein are shown by yellow circles; the mass calculated from these peaks is shown above in Da.
B- A mass spectrum of the same protein, alpha synuclein, measured directly from human cells after the protein was introduced into the cell. The mass now differs slightly due to modifications that occur in human cells.
C- The mass spectrum of another protein, Bruton’s tyrosine kinase, which was produced genetically in the cells. This protein also has modifications that are typical of human cells.
This is important both to making the lives of researchers easier, but also because proteins are produced for use in many biotech industries, and native-MS is an excellent method for quality control of such proteins directly from the production facility. Moreover, by examining proteins and protein complexes directly from the cell instead of after purification, there is an opportunity to capture interactions reflective of the protein’s role in the cell.
The Sharon lab started using direct-MS to study proteins produced in bacterial cells1. In initial studies, the Sharon lab demonstrated that a full suite of native-MS experiments is feasible without the need for protein purification. Elegant work from colleagues in the lab has shown that direct-MS can quantify how different mutations affect the energetics of protein-protein interactions 2,3. Direct-MS can also be used to study proteins produced in eukaryotic cells and exported out of the cell4,5. For example, antibodies, important immune proteins that are widely used in biochemistry, can be measured from crude samples. Native-MS can then report on whether these antibodies are modified in the cell by the addition of sugar molecules. We are also able to compare the stability of different antibodies, which is an important property in the production of these antibodies for therapeutic proteins.
My current research focuses on applying direct-MS to proteins produced in human cells that are not exported out of the cell. When a human protein is produced in a human cell, it is most similar to the biologically relevant version of that protein, since the human cell modifies it in unique ways. By developing new protocols for producing proteins and preparing samples, I am enabling the lab to easily use native-MS to study the full range of proteins that interest a molecular biologist. I have applied two different methods for getting the proteins into the human cells. I can either introduce an already produced protein into the cell by forcing it to cross the cell membrane, or I can give the cells pieces of DNA that will induce them to produce the protein themselves. I have shown that both methods enable us to detect a range of proteins directly from the crude lysate of human cells, and that these proteins are modified by human cells in ways that they would not be modified in bacterial cells (see Figure 1). I am now working to apply my protocols to understanding the range of interactions that we can measure and quantify for a given protein using the direct-MS approach.
On any given day, I can be found working with cells to prepare direct-MS samples or working at the mass spectrometer studying the proteins found in those cells. Yet I never cease to be amazed at the richness of information that I can extract from the simple act of measuring mass.

Articles are written by Fulbright grantees and do not reflect the opinions of the Fulbright Commission, the grantees’ host institutions, or the U.S. Department of State.


